Ryan Sullivan1, Haofei Feng2, Vipul Patel3, Murray-Luke Peard4, Chris Albone5
1University of Sydney, Sydney, ryan.sullivan@sydney.edu.au
2University of Sydney, Sydney, haofei.feng@sydney.edu.au
3University of Sydney, Sydney, vipul.patel@sydney.edu.au
4University of Sydney, Sydney, murray-luke.peard@sydney.edu.au
5University of Sydney, Sydney, chris.albone@sydney.edu.au
Introduction
Imaging research, clinical, preclinical, or otherwise, is often multisite, multimodal, and compute intensive. XNAT is an imaging data platform that has been rapidly gaining popularity both worldwide and throughout Australian research institutions and facilities [1]. As part of the University of Sydney’s Core Research Facility program, we have developed our Imaging Data Service (IDS) using XNAT as one of the core technologies. IDS is able to ingest, store, and analyse data in an automated and compliant manner to facilitate clinical workflows.
We have connected instruments in the Sydney Imaging Core Facility and I-Med, a local clinical site. We will be expanding to cover instruments in three schools along with additional clinical sites over the coming year. We will discuss challenges we’ve encountered in terms of developing these systems, as well as hurdles in dealing with patient privacy and vendor software.
PROCESS
Acquired images are passed directly from equipment to a Clinical Trials Processor (CTP) or Research Automated Project Allocator & Anonymiser (RAPPA) on site, where direct patient identifiers are stripped in a compliant manner before the data is sent to the XNAT repository. The direct identifiers are stored on-site in such a way that allows automated re-association of derivative data and analysis results on site to facilitate clinical workflows. Other patient data not captured at the instrument are stored in a separate REDCap system, linked with a common anonymised key. This allows a higher granularity of control to address the different needs of a variety of projects and sites based on patient consent. Data from other repositories, such as historical data on our Research Data Share (RDS), may also be batch uploaded to the new system.
Once stored in XNAT, researchers may access their data using AAF authentication via web browser or though multiple clients and connected platforms using the REST API. We have implemented XNAT’s pipeline engine using containerised workflows run on Artemis, our HPC, as the backend, with the future aim of being able to run on private and public clouds. Alternatively, they may use resources such as Argus, Sydney’s Virtual Research Desktop, or the ARDC’s curated Characterization Virtual Lab (CVL). Finally, we look at integration with two informatics platforms, Jupyter Hub and Nipype, through which workflows may be developed. This gives researchers the freedom to choose the desired technologies for their particular workflows.
From the user’s perspective, this provides a “big green button” solution to analysing their data using tested and curated pipelines, while also providing tools for power users who wish to delve deeper into informatics development.
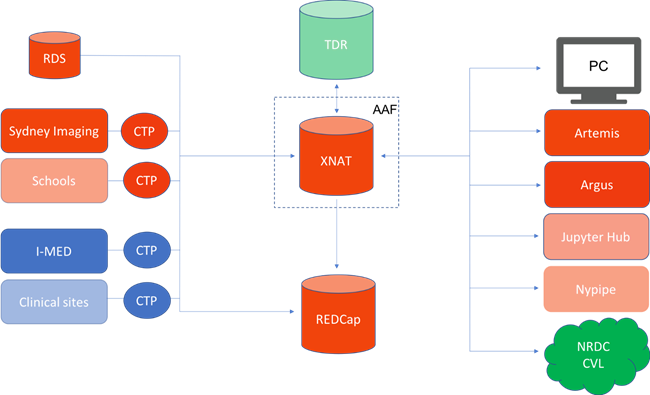
Figure 1: High level overview of data flow in our Imaging Data Service. Orange are systems belonging to the University of Sydney directly, Blue belong to partner institutions, Green belong to NCRIS capabilities. Partially transparent items are planned over the coming year, but not yet in production.
FUTURE WORK
We continue to look at developing sustainable DevOps frameworks for pipelines to allow the system to be self-sustaining and get ICT “out of the way.” Next steps are continued rollout to appropriate faculties, improved auditing and reporting frameworks for research integrity, operations ROI, and data provenance. We are also interested in discussing interfacing with other similar systems meeting the TDR standard.
REFERENCES
- Marcus, D. S., Olsen, T. R., Ramaratnam, M., Buckner, R. L., The extensible neuroimaging archive toolkit. Neuroinformatics, 2007. DOI: 10.1385/NI:5:1:11
Biography:
Dr Sullivan is a biophysicist with an interest in neural implants. His research led him into software development for automatic characterization of implants and neural tissue. Dr Sullivan joined the University of Sydney in 2017 where he now works on eResearch projects focusing on characterization domains.